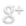Ligation reaction:
x ul DNA insert
y ul DNA vector
2 ul 10x T4 Ligase Buffer (make sure it is well dissolved)
z ul H2O to 20 ul total
1 ul T4 DNA Ligase (400U/ul) very temperature sensitive (keep cold at
all times)
10 minutes at RT (20-25C), then put on ice for transformation
y ul of vector = (t * SizeVec/SizeIns) / (3 + SizeVec/SizeIns)) / [vector]
z ul of water = 20 – x – y

[python filename=”ligation2.py”]

in ng, and then use the known concentration of X/ul and Y/ul to
calculate volumes x and y.
Want to use 20-200ng total DNA in rxn, aim for 100ng first if possible
v = ng of vector (mass)
i +v = t , where t=100 ng, t is total mass (equation 1)
v = moles vector * MWvector (g/mol) * 109 ng/1g
MW is proportional to size
i = 3* v (SizeIns/SizeVec) (equation 3)
solving for i.
i * SizeVec/SizeIns = 3t – 3i
i * SizeVec/SizeIns + 3i = 3t
i * (SizeVec/SizeIns +3) = 3t
i = 3t/ (SizeVec/SizeIns +3) (equation 4)
v= (t * SizeVec/SizeIns) / (3 + SizeVec/SizeIns) (equation 5)
v= 100 * SizeVec/SizeIns) / (3 + SizeVec/SizeIns) (equation 5a)
Practical equations:
x ul of insert = (300/ (SizeVec/SizeIns +3)) / insert [concentration](equation 6)
x ul of insert = (3t/ (SizeVec/SizeIns +3)) / insert [concentration] (equation 6a, general version, where t = total DNA)
y ul of vector = (100 * SizeVec/SizeIns) / (3 + SizeVec/SizeIns)) /vector [concentration] (equation 7)
y ul of vector = (t * SizeVec/SizeIns) / (3 + SizeVec/SizeIns)) /vector [concentration] (equation 7a, general version, where t = total DNA)
z ul of water = 20 – x – y (equation 8)
(equation 8)
equations 3, 4 and 5 (example t = 20 ng)