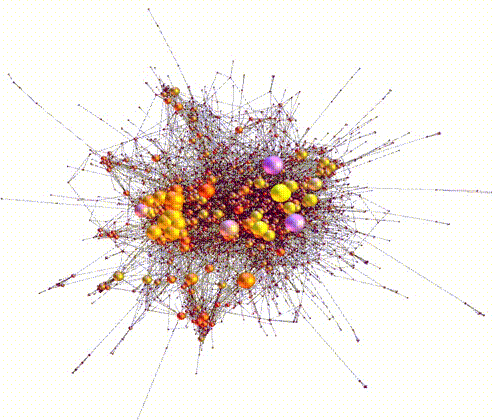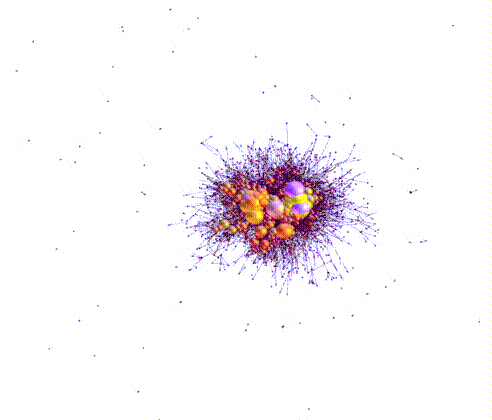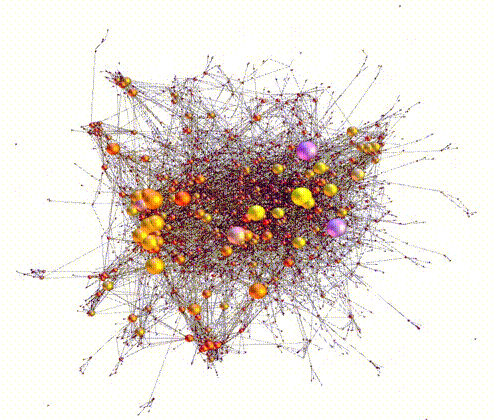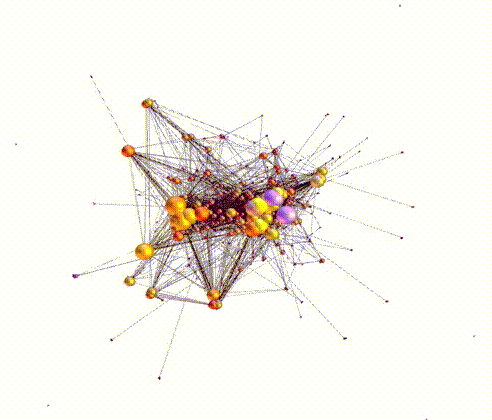
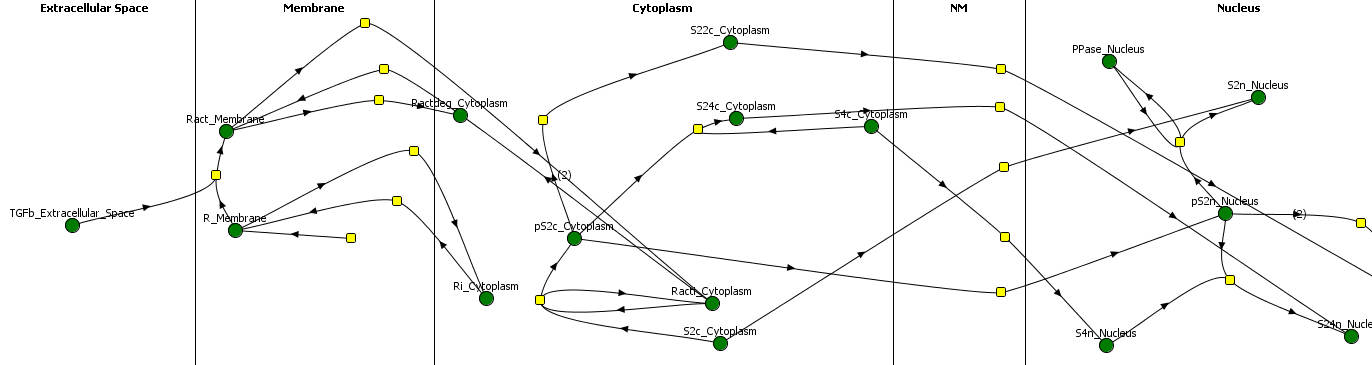
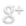Large sets of “Omics” data, for example, genomic, transcriptomic, epigenomic, etc, have powered the burgeoning discipline of systems biology. Systems biology is the study of biological processes within a cell or organism organized as a hierarchy of systems, usually visualized in terms of networks and subnetworks. The success of synthetic biology/biosystems engineering is in no small part due to advances in systems and computational biology.
Systems and computational biology research at RSI focuses on developing dynamic models of evolving systems. Key to our work was the development of the first dynamic model of a living organisms, mycoplasma genitalium by Markus Covert’s group at Stanford, which can be accessed at wholecellviz (http://wholecellviz.stanford.edu/). We are using his model as a framework to develop models that evolve by introducing biologically plausible mutations under various selections. RSI is developing the computational infrastructure and algorithms necessary to simulate cells and organisms
One of the greatest challenges to human health and overcoming aging is cancer. The primary problem with developing therapies is that cancer cells are the ultimate evolvers. Freed from the constraints necessary for organismal homeostasis and reproduction, the evolutionary constraints on cancer cells are far less than on normal organisms, and one of the key changes in many cancers are significantly increased rates of mutation and genetic recombination due to chromosomal instability. Most single drug therapies for cancer, in which large pharmaceutical companies are deeply invested, are doomed to failure because the population of cancer cells evolve resistance by multiple mechanisms: not only gain and loss of function point mutations, but creation of entirely new functions by creation of fusion proteins such as BCR-ABL. Even rational therapies that might strike at the key aspects of cancer cell function, such as rapid cell division, or telomere elongation can be overcome by mutations altering drug transport or activation of new pathways, such as ALT. However, there are some constraints on cancer evolution. At RSI, we seek to identify potential evolutionary constraints, especially context-dependent constraints based on the current state and history of the cancer population to predict the future trajectory of change. With such a trajectories, it will become possible to design more effective drugs and to combine existing drugs to suppress cancer. The identification of key biomarkers in the context of evolutionary trajectories is ongoing at RSI
However, ultimately it may be that the oldest treatment for cancer, surgery, will prove to be most effective. How can surgery address distant metastases? Conventional surgery cannot, but microsurgery performed by biological robots very well might.
Systems biology of regeneration for regeneration engineering
To understand the differences between organisms with super regenerative powers, such as planaria and hydra, moderate regenerative power, such as newts, and very modest regenerative power, such as humans; RSI is studying and comparing the architecture of the transcriptomic and epigenomic networks that underlie stem and differentiated cell function. We are especially interested in understanding how the neoblasts recognize positional information in the planarian and have established programs to recreate biological structures without recapitulating early development. Elucidation of regulatory networks and comparison with IPS, ES and adult stem cells in humans and mice are of great importance.
Systems biology of aging
RSI is developing computational models of human and mammalian aging using deep learning based on modeling instability of the differentiated state with time using available transcriptomic, epigenomic and proteomic data. Epigenetic drift, first identified by Stuart Kim at Stanford in the nematode C. elegans, is a key feature of aging in animals. Changes in the epigenome, resulting in loss of information, are probably partially responsible for gradual loss of differentiated cell function, which may follow a series of distinct steps. Identification of these steps, which may be species specific, may allow direct intervention to slow or reverse key aging phenotypes for extended periods of time until a subsequent change requires further intervention. This intermediate strategy may have substantial health benefits for humans.
.