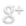iGEM 2007 Silver Medalists and Grand Prize Finalist + Best Poster
Our team consists of students from University of California at Berkeley and Stanford University. Our goal is to repair damaged hearts. We are engineering embryonic stem-like cells to autonomously differentiate into cardiomyocytes, recognize and bind to damaged heart cells and then activate a defined program. Bound cells will express anti-apoptosis and anti-necrosis proteins to prevent cell death, express increased amounts of a cell-cell adhesion molecule to stimulate the formation of a sheet of cells and to cause some of the cells to penetrate into the inner layers of the heart. Cells will also express a growth factor to stimulate new blood vessel formation. Cells that fail to bind in a defined amount of time are programmed to self-destruct.
Repairing Infarcted Hearts
Goals
The problem: 1,200,000 people suffer a (new or recurrent) MI every year, and about 40% of them die as a result of the attack. Current therapies are inadequate. Drugs at best can only stabilize the condition. Endogenous adult atem cells do not repair the heart, and the heart does not regenerate.
Our goal is to repair damaged hearts. We are engineering embryonic stem-like cells to autonomously differentiate into cardiomyocytes, recognize and bind to damaged heart cells and then activate a defined program. Bound cells will express anti-apoptosis and anti-necrosis proteins to prevent cell death, express increased amounts of a cell-cell adhesion molecule to stimulate the formation of a sheet of cells and to cause some of the cells to penetrate into the inner layers of the heart. Cells will also express a growth factor to stimulate new blood vessel formation. Cells that fail to bind in a defined amount of time are programmed to self-destruct.
Autonomous Differentiation
Since differentiation of P19 Cells into Cardiomyocytes takes 8-12 days, we will begin differentiation before injection. This means we can do things in vitro. We will differentiate P19 cells using BMP4 and transcription factor Nkx2.5 and then use G418 resistance (http://www.invivogen.com/family.php?ID=6) to select for those cells that differentiated into cardiomyocytes. BMP4 should bypass the requirement for cellular aggregation in cardiomyocyte differentiation. In addition, we will add parallel circuits with GATA4 and MEF2C, both of which will hopefully enhance differentiation.
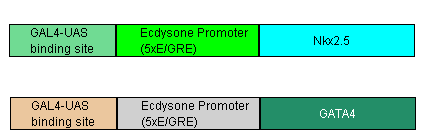
- Expression of BMP4
- Next, we will use an Nkx-dependent promoter to trigger the BMP4. We will also add an enhancer (such as GAL-4) to ensure that once BMP4 has been switched on, it will stay on.
- The Nkx-dependent promoter will include the Nkx2-5 binding site 5′-TNAAGTG-3′, as well as a minimal promoter such as Cmv or SV40.
- In addition to BMP4 we will also have parallel circuits (using the same Nkx promoter) for both GATA4 and MEF2C. These transcription factors will hopefully enhance differentiation success rates.
- Next, we will use an Nkx-dependent promoter to trigger the BMP4. We will also add an enhancer (such as GAL-4) to ensure that once BMP4 has been switched on, it will stay on.
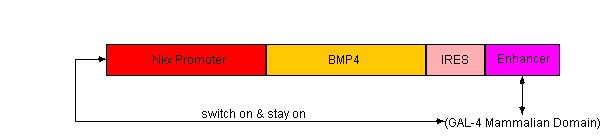
- G418 Selection
- We will create a fusion gene consisting of alpha Cardiac MHC promoter and aminoglycoside phosphotransferase. Since Alpha-MHC is expressed in differentiated cardiac tissue, we will use antibiotic resistance to select only the differentiated cardiomyocytes.
-
- BsrGI and BsaHI will be the restriction enzymes used to cut out the alpha Cardiac MHC promoter sequence from chromosomal murine DNA. NeoR will be cut out from a plasmid containing the resistance gene and then fused with the alpha Cardiac MHC promoter.
Timed Stem Cell Self-destruction
Construct 1
Construct 2
Construct 3
Construct 4
Construct 5
Rescue Design
Switch OFF
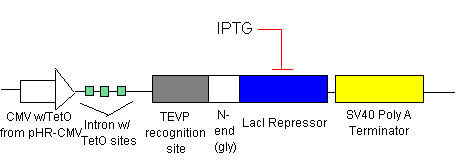
- Initially, there will be no IPTG added.
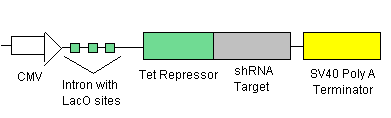
- LacI Repressor (see line 1) will be inhibiting transcription at the introns with the LacI Operator sites (see lines 2 &3). This means that there is no TRXregI Repressor, TEVP, lambda activator and thus no BAX being created.
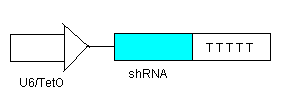
- To ensure that there is no leakage in the system in the off state, shRNA (see line 1) will be inhibiting transcription of the elements before each shRNA target site.
Switch ON
- To turn on the system, we will add just enough IPTG (which inhibits the LacI Repressor) to decrease the LacI Repressor concentration on the introns so that lines 2 & 3 begin to be transcribed. The production of TEVP will create a positive feedback loop on itself as it cuts the proceeding LacI Repressors for degradation. It will also cut the LacI Repressors inhibiting the TRXregI Repressor site, allowing for some TRXregI to be produced. This will repress the TRXregI Operator site thus preventing the production of both LacI Repressor as well as shRNA.
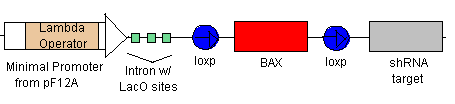
- Because of the positive feedback loop, TEVP will gradually increase in concentration along with the Lambda Activator. The Lambda Activator bind to the Lamda-vp16 Operator in the Minimal Promoter. Because of to cooperative binding, BAX will begin to be transcribed only when the concentration at the Operator reaches the Kd. BAX will eventually reach the threshold levels and cause the cell to apoptose.
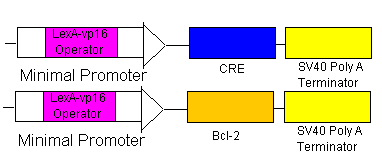
Rescue
- If the rescue signal LexA-vp16 is received from Omar/Hugo, then it will bind to the minimal promoters (see lines 5 & 6) which will begin the production of CRE (which will flip the BAK at the loxp sites) and also begin the production of Bcl-2 to combat the already produced BAX. Yay! Salvation!
Structure inspired by: Dean et al,.(2007) A Tunable Genetic Switch Based on RNAi and Repressor Proteins for Regulating Gene Expression in Mammalian Cells
Activation Of Stem Cell Effectors
CRP-based Targeting and Signaling
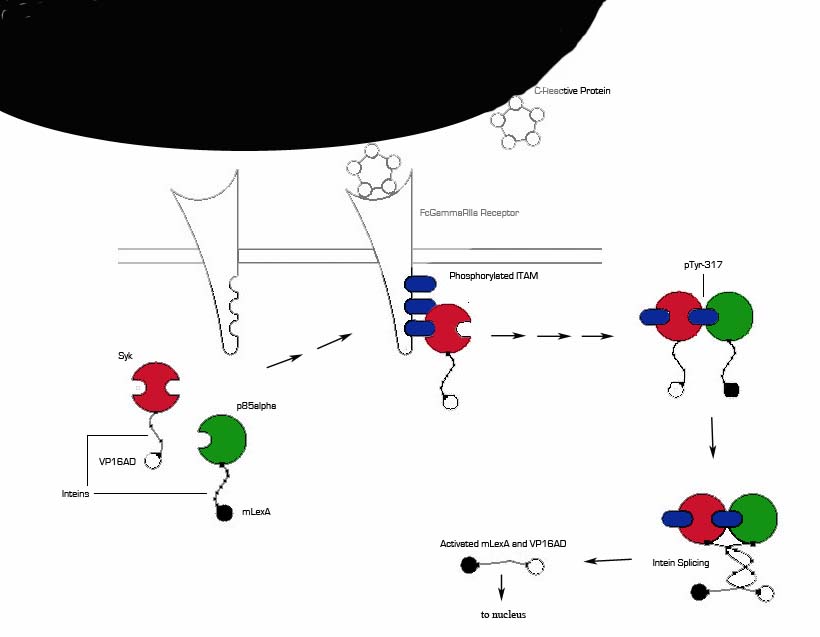
1) MI induces large increase in serum CRP levels. It also induces CRP expression in cardiomyocytes.
2) CRP deposits onto damaged myocardial cells (it binds to exposed small nuclear ribonucleoproteins on dying cells)
3) Our cells expressing FcGammaRIIa-R131 bind CRP on the surface of damaged cells and in the serum surrounding the damaged area.
4) Crosslinking of the receptors induces the phosphorylation of the ITAMs in the intracellular region of the receptor.
5) Syk is recruited to the receptor and becomes phosphorylated at ITAMs.
6) Crosslinking of the receptors also recruits p85 which binds to the phosphorylated Tyr-317 residue on Syk.
7) This interchange allows for the inteins (dnaEc-VP16AD at C terminus of Syk and dnaEn-mLexA at N terminus of p85) to interact and self-splice.
8) Self splicing of the inteins produces the VP16AD-mLexA needed for downstream signaling.
Benefits: Binds to CRP on damaged cells to allow for targeting of therapeutic agents (versatile since CRP is expressed in a variety of inflammed tissues). Our system also removes CRP from the region surrounding the damaged area, lessening the damage caused by CRP and increasing the chance of full recovery.
Problems: FcGamma can bind IgG present in serum. Although the FcGammaRIIa-R131 exhibits weak affinity for IgG the possibility of binding and crosslinking the receptor is still there. This problem can be alleviated if the the FcGamma-CRP system is used as part of an AND gate with another receptor system.
CRP-based Targeting and Signaling
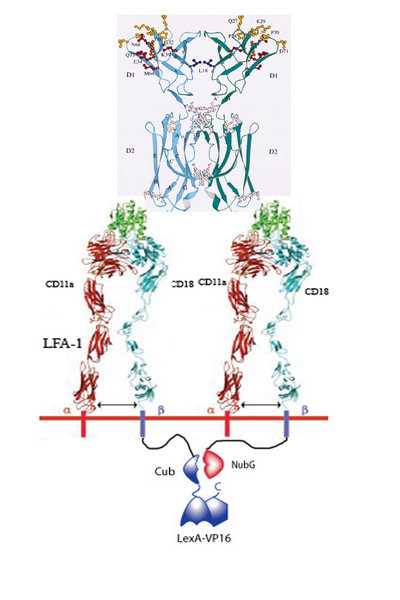
1. Infarced myocardium induces expression of ICAM.
2. Two LFA-1’s on our programmed cells bind to the same ICAM dimer on a damaged cardiomyocyte.
3. 50% of the time LFA-1 – Cub will be in close proximity of LFA-1 – NubG.
4. When this occurs, the Cub and NubG of split ubiquitin interact and reconstitute the ubiquitin.
5. Ubiquitin protease then cleaves the Cub portion of ubiquitin and releases the transcription factor LexA-VP16.
6. LexA-VP16 travels to the nucleus, binds to the lexA operator minimal promoter, and effectors such as N-cadherin, VEGF, are upregulated.
Anti-myosin Heavy Chain Targeting And Signaling
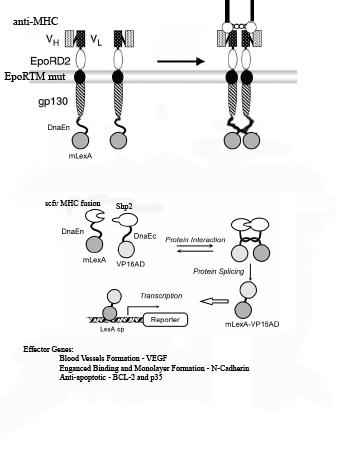
1) Myosin Heavy Chain (MHC) is exposed on infarcted heart tissue.
2) scFv chimeric receptor targeting MHC, binds to a repeated domain of MHC.
The scFv chimeric receptor contains a fusion of the following protein domains: anti-MHC-Fc, EporD2, EporTM, and gp130i, dnaEC-VP16. EpoR= erythopoeitin receptor. dnaEC= carboxy domain of the Sp Synechosystis split intein.
3) EporD2 facilitates the dimerization of the chimeric receptors
4) A mutant version of EporTM inhibits ligand independent dimerization of the chimeric receptors
5) gp130i becomes activated and JAK’s phosphorylate tyrosines on gp130i, including Y759.
6) Shp2-mLexA-dnEN binds to phosphorylated Y759 of gp130i. (mLexA is a mutated form of E coli DNA binding protein LexA that does not contain a cryptic nuclear localization sequence found in the wild-type).
7) Shp2-mLexA-dnEN comes in close proximity to the dnaEC-VP16 fused to the c-terminus of the gp130i
8) dnaEN and dnaEC, parts of the Sp Synechosystis split intein system, undergo peptide processing resulting in the release of newly constituted fusion transcription factor mLexA-VP16.
9) mLexA-VP16 translocates to the nucleus where it binds to minimal promoters containing lexA operators to switch on expression of cell-cell interaction proteins such as N-Cadherin , anti-apoptotic proteins such as Bcl-XL,p35 and IAP and blood vessel growth factors such as VEGF.
Cell Integration
Increased levels or strength of adherins will cause cells to penetrate into tissue with lower levels of cell-cell interacting molecules. Activation of signaling by binding of one of our signaling targeting chimeric proteins will trigger expression of the cell-cell adhesion molecule N-Cadherin which was inserted downstream of a minimal promoter with lexA operators. Increased N-Cadherin expression may result in penetration of the new cardiomyocytes into the myocardium.
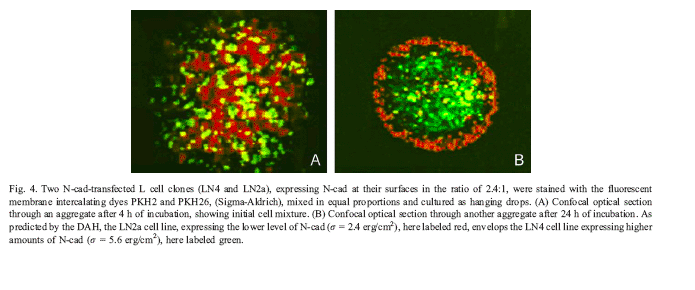
Ilustrative data demonstrating the basic principle from the literature:
Rapid Assembly of Biobrick Parts
Because of the need to rapidly create our constructs (we started building constructs in the middle of August), the incompatibility of many of our sequences with standard Biobrick restriction enzyme sites, and the relative dearth of relevant Biobrick parts, we used PCR overlap assembly technology to assemble our parts all at once, instead of using the stepwise Biobrick method.
The Bay Area RSI Team
Regenerative Sciences Institute is proud to field the Bay Area RSI Team. Our team consists of Berkeley and Stanford students who have come together to work at RSI.
Our team members:
Molly Allen, UC Berkeley
Ed Cho, Stanford University
Hugo Decker, Stanford University
LeAnn Duong, Stanford University
Chian Gong, UC Berkeley
Omar Kahn, UC Berkeley
Jennifer Lei, UC Berkeley
Anastasia Shabalova, UC Berkeley
Jack Tung, UC Berkeley
Our instructor: Andrew Mendelsohn, PhD, RSI, Instructor
How It Worked
Our project started late at the end of June. After an introduction to molecular and synthetic biology, the students decided on a project which would involve cell self-assembly and solving a real-world problem. The students decided to try to repair damaged heart tissue by engineering cells to differentiate and target damaged myocardium. The students split into 3 teams: Timed ES cell Self-Destruction, Differentiation, and Targeting/Cell Signaling. The students designed the overall approach of each team, and the specific circuits and oligo design themselves. Dr Mendelsohn provided useful critical input and suggestions to help guide the teams, but the teams had ultimate responsibility for the projects.
Status
Because of our late start, the Timed ES cell Self-Destruction and Differentiation teams only made modest progress. However, the Targeting/Cell Signaling team was able to continue working and have just (10/26) built the CRP signaling circuit and are very close to completing the ICAM and scFv circuits.
Thanks!
RSI and the Bay Area RSI Team wish to thank the Abbott Foundation and Panorama Research for their support.